scMultiNODE: Integrative and Scalable Framework for Multi-Modal Temporal Single-Cell Data
 Visualization of scMultiNODE
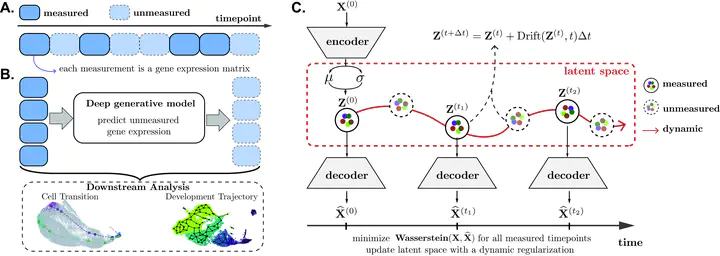
Visualization of scMultiNODEWe introduce scMultiNODE, an unsupervised integration model that combines gene expression and chromatin accessibility measurements in developing single cells, while preserving cell type variations and cellular dynamics. First, scMultiNODE uses a scalable, Quantized Gromov-Wasserstein optimal transport to align a large number of cells across different measurements. Next, it utilizes neural ordinary differential equations to explicitly model cell development with a regularization term to learn a dynamic latent space.